Tropical Medicine Research Center for Talaromycosis in Vietnam

Principal Investigator: Thuy Le
Lead Institution: Duke University, USA
Background
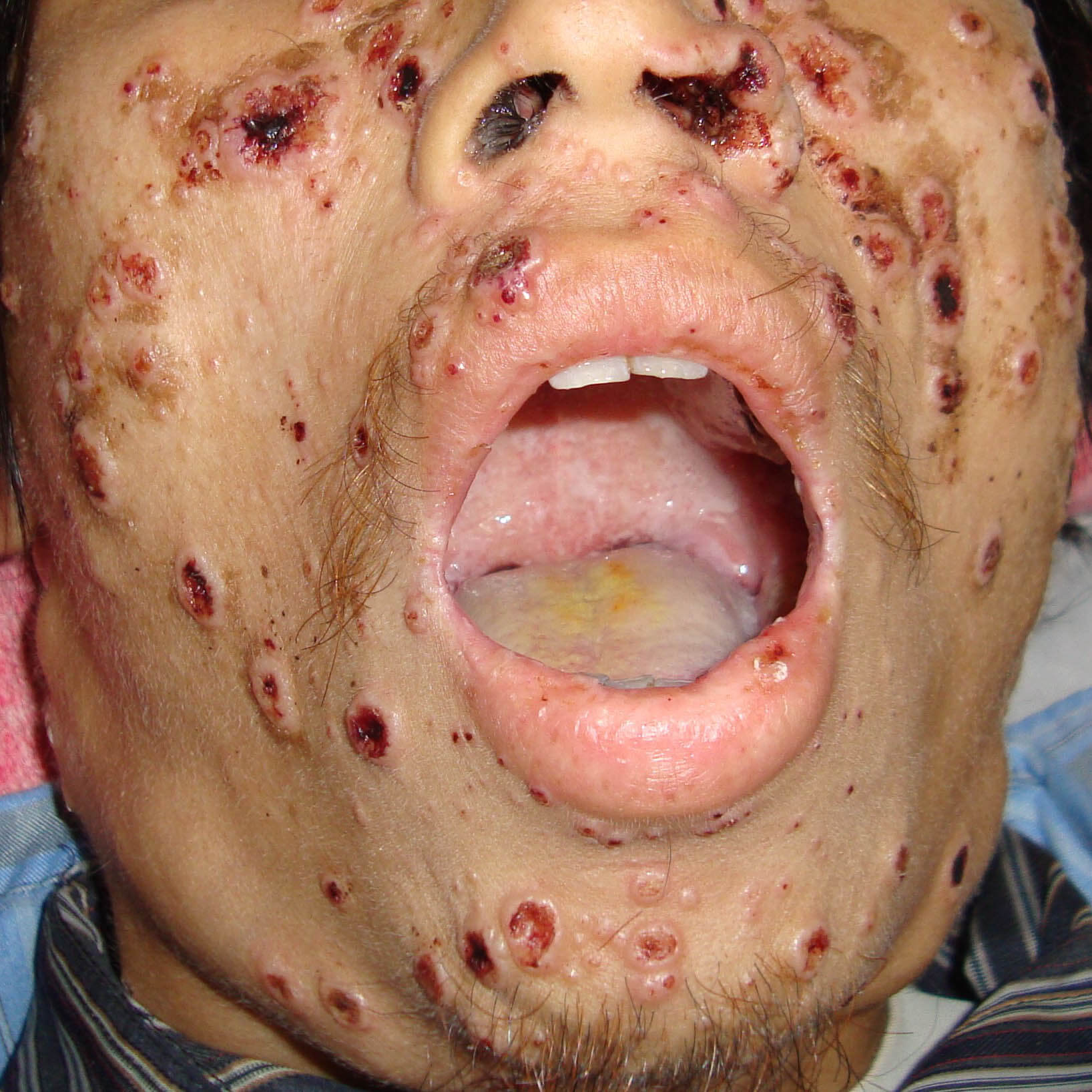
Talaromycosis (formerly penicilliosis) is a life-threatening invasive fungal infection caused by the dimorphic fungus Talaromyces marneffei (Tm) that is endemic in countries of Southeast Asia and is one of seven endemic mycoses of medical importance. Even with antifungal therapy, one in three patients die because the diagnosis is often made too late for the antifungal drugs to work. Impediment to our progress is our reliance on decade-old culture-based diagnostic methods which take up to 28 days for identification and are poorly sensitive.
Scientific Aims
- To validate a pipeline of novel pathogen-based diagnostics for talaromycosis that i) overcome many limitations of the current culture methods, ii) are in late stages of development, and iii) have the potential to be scaled up and commercialized for clinical and public health applications.
- To determine the differential host transcriptional signatures that differentiate talaromycosis from other opportunistic infections.
- To conduct the first-ever community-based seroprevalence and air and soil sampling study to determine the link between airborne exposure and highland geography to human disease.
Hypotheses
We hypothesize that factors at each step of the parasite life cycle interact to determine morbidity in schistosomiasis; in particular, that contributors to greater disease severity in LA compared to LV include:
- The Tm qPCR and 3 antigen-detection assays will be at least 90% sensitive and 96% specific in the detection of talaromycosis
- Whole blood RNA expression profiles are distinct in patients infected with Tm vs. TB and other opportunistic infections and can be used to identify a diagnostic gene set for talaromycosis.
- Anti-Tm IgG seroprevalence in humans is correlated with Tm burden in the air and is at least two times higher in highland versus lowland regions.
Design
- We will validate 4 novel pathogen-based assays for Tm diagnosis – a real-time PCR assay, a Wantai Mp1p enzyme immunoassay, a D41 lateral flow antigen assay, and IMMY Mp1p lateral flow antigen assay - using whole blood, sera, and urine samples being collected as part of ongoing NIH-funded talaromycosis screening cohorts. We will also validate the 3 antigen assays in a cohort of non-HIV infected patients with talaromycosis at Guangxi Univ in China.
- We will perform RNA sequencing of blood samples at enrollment from a subset of patients with and without Tm to identify Tm-specific gene sets, which will be validated by qRT-PCR using samples from another subset of patients with and without Tm.
- We will conduct an integrated community-based epidemiological study that combines blood sampling of agricultural/animal farm workers and their respective environmental samples of air and soil in the highland vs. lowland provinces. Sampling of soil and air in each province will be from the same location where the human volunteers are recruited and will be during the same time window.